Bioimage analysis with Napari
I have just completed my lecture notes on Bioimage Analysis with Napari, which I have developed for our Master of Neuroscience programꜛ this year. I avoided using any copyright-protected material so I could make the entire lecture freely available. It is available under Teaching, where you can find other free resources as well. The course is specifically designed to help unlocking the potential of Napari – without requiring any programming skills in Python. The focus lies on its practical usage and the material is aimed at a general audience, including anyone eager to learn about bioimage analysis and to get started with Napari.
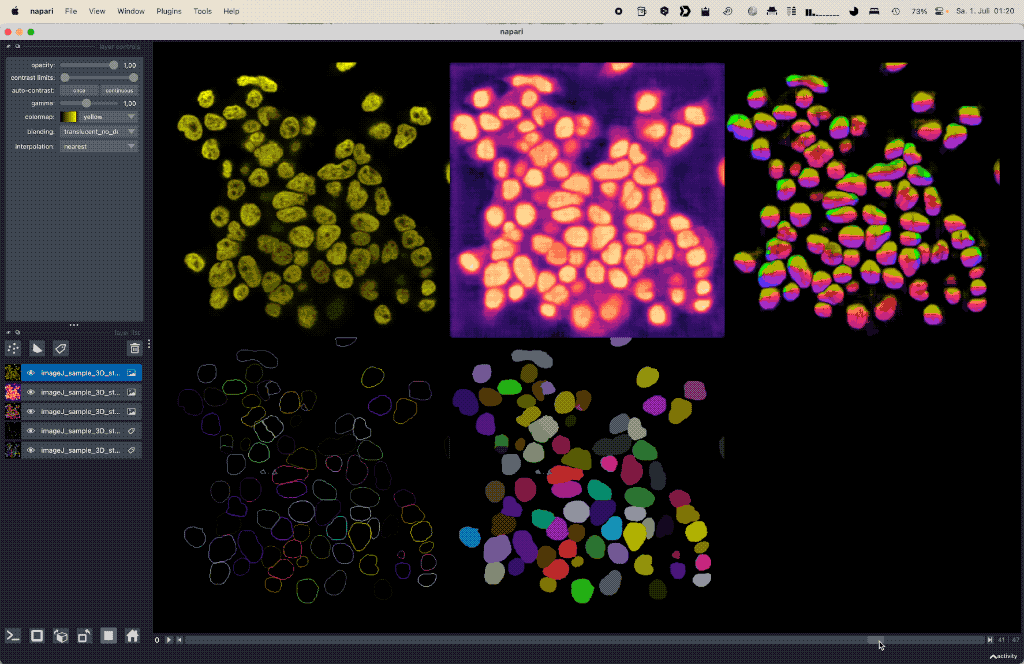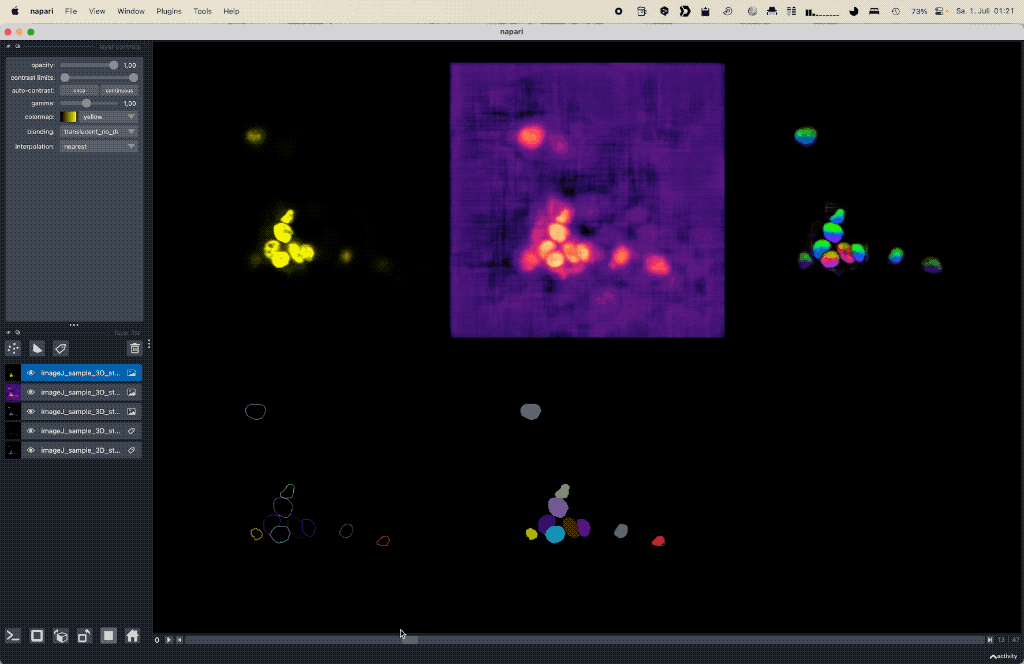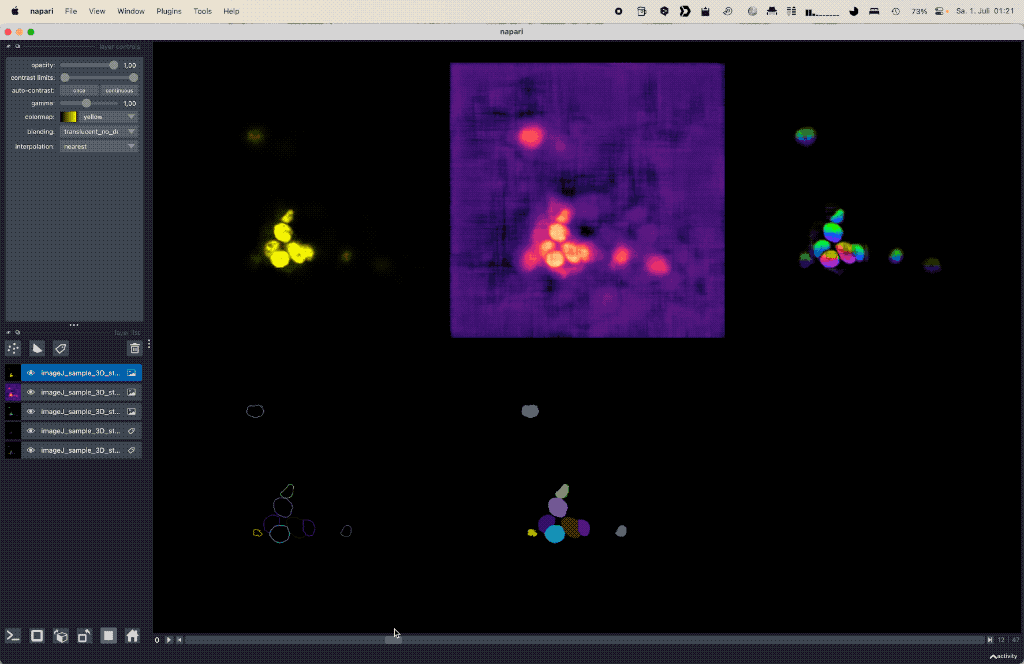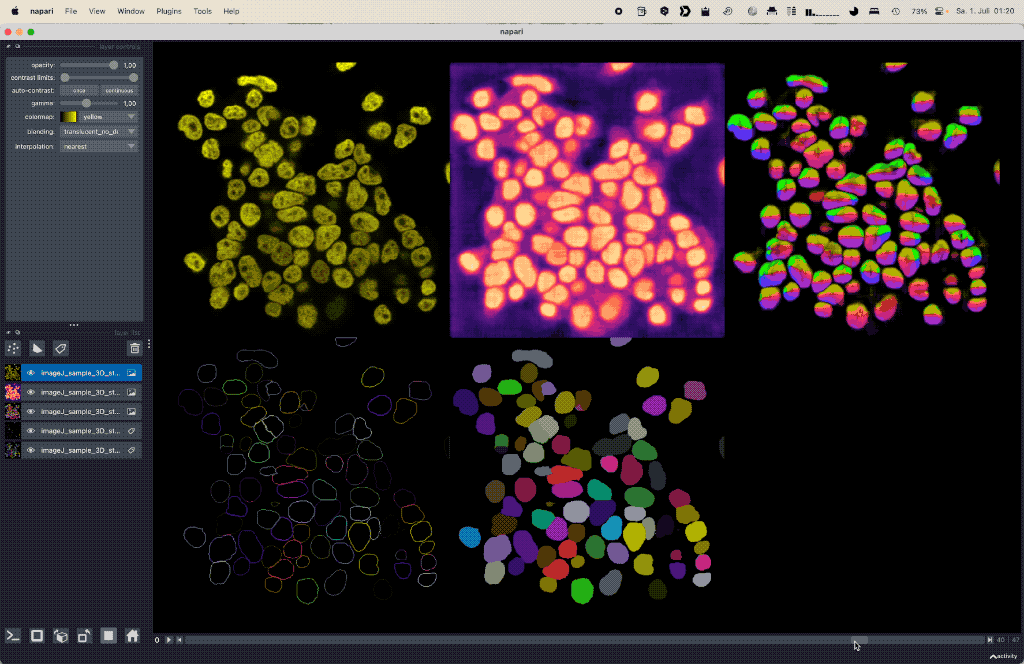 Using deep learning based Cellpose for versatile semantic cell segmentation.
Using deep learning based Cellpose for versatile semantic cell segmentation.
What does the material contain?
The material consists of short tutorials that cover the essentials of working with Napari. You will find step-by-step guidance on the installation of Napari, basic handling, common image processing techniques such as image denoising, and delve into advanced methods like deep-learning based imaging segmentation, cell tracking, cell colocalization, and filament tracing. My goal was to make this resource comprehensive yet approachable, ensuring that you can easily grasp the practical use of Napari in common bioimage analysis workflows. Each tutorial is self-explanatory, thus, can be used as a stand-alone resource for self-study. I have also included a list of additional resources for those who wish to explore each of the topics further.
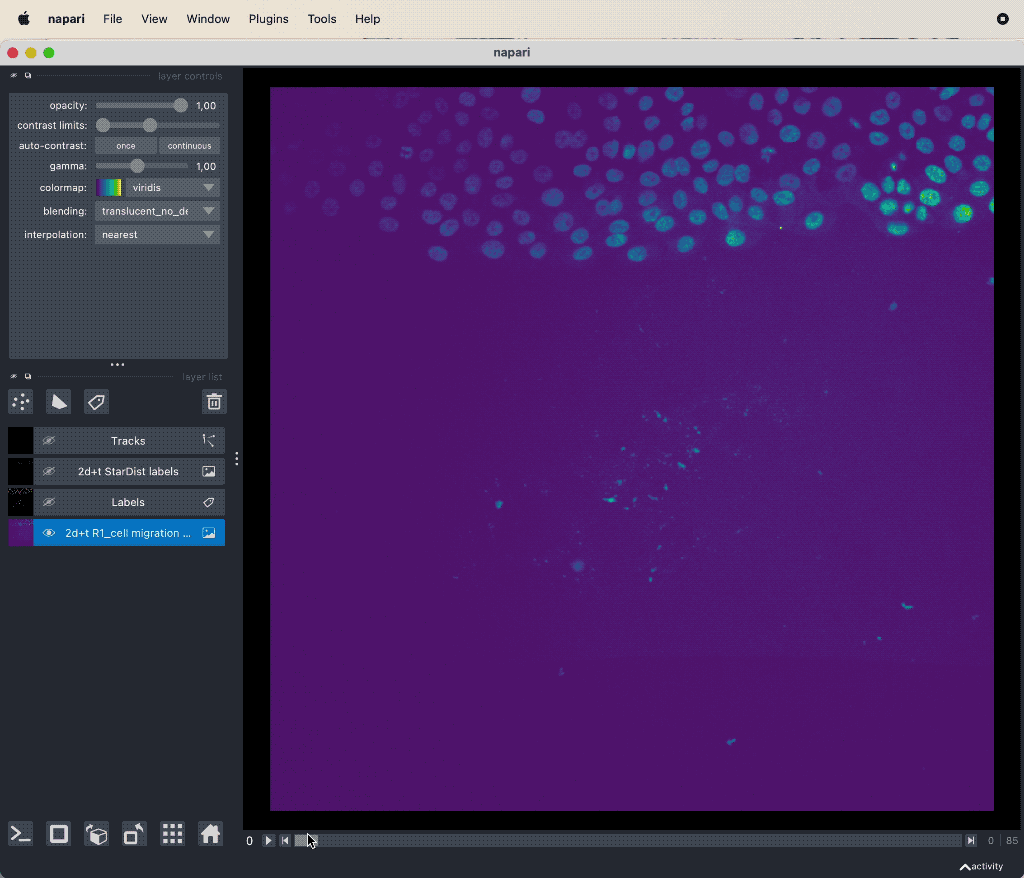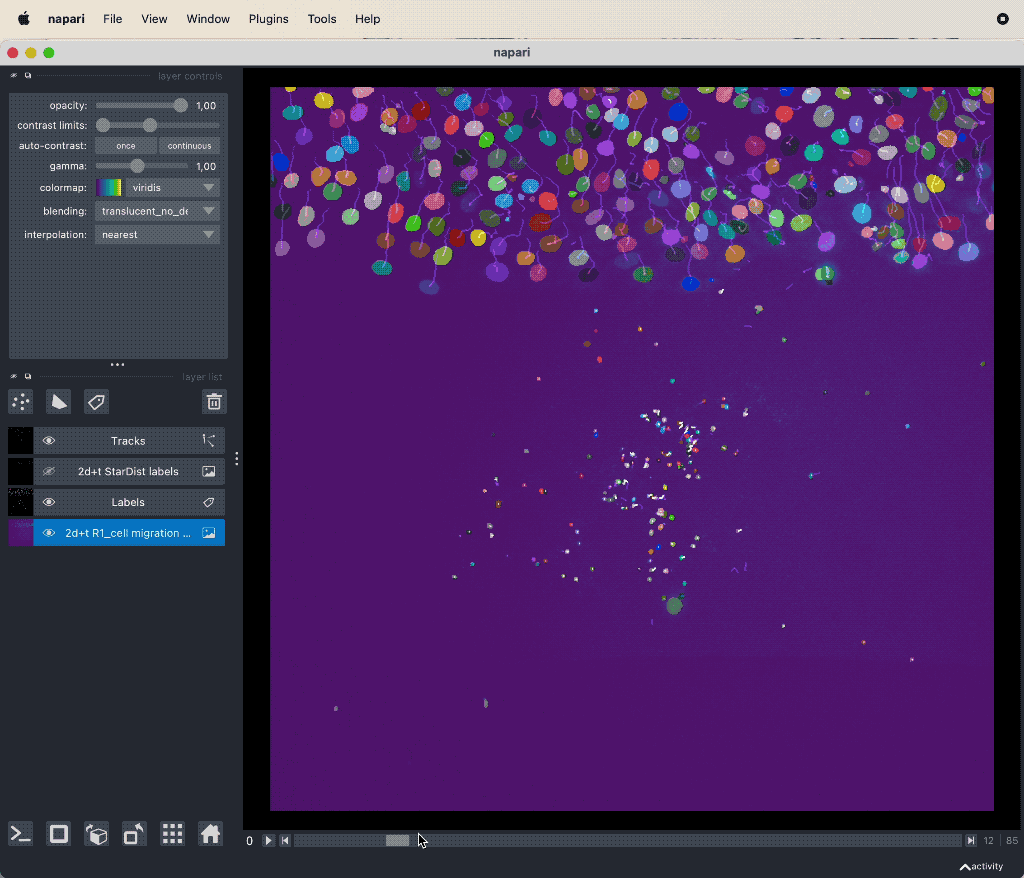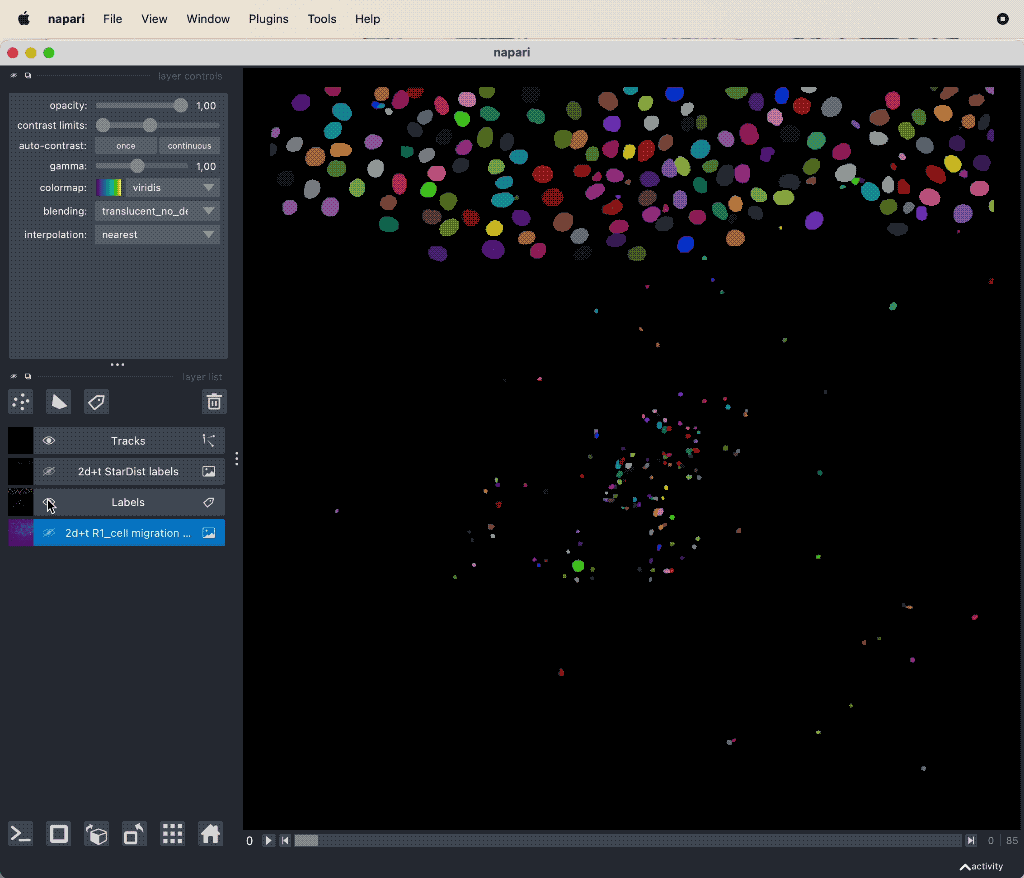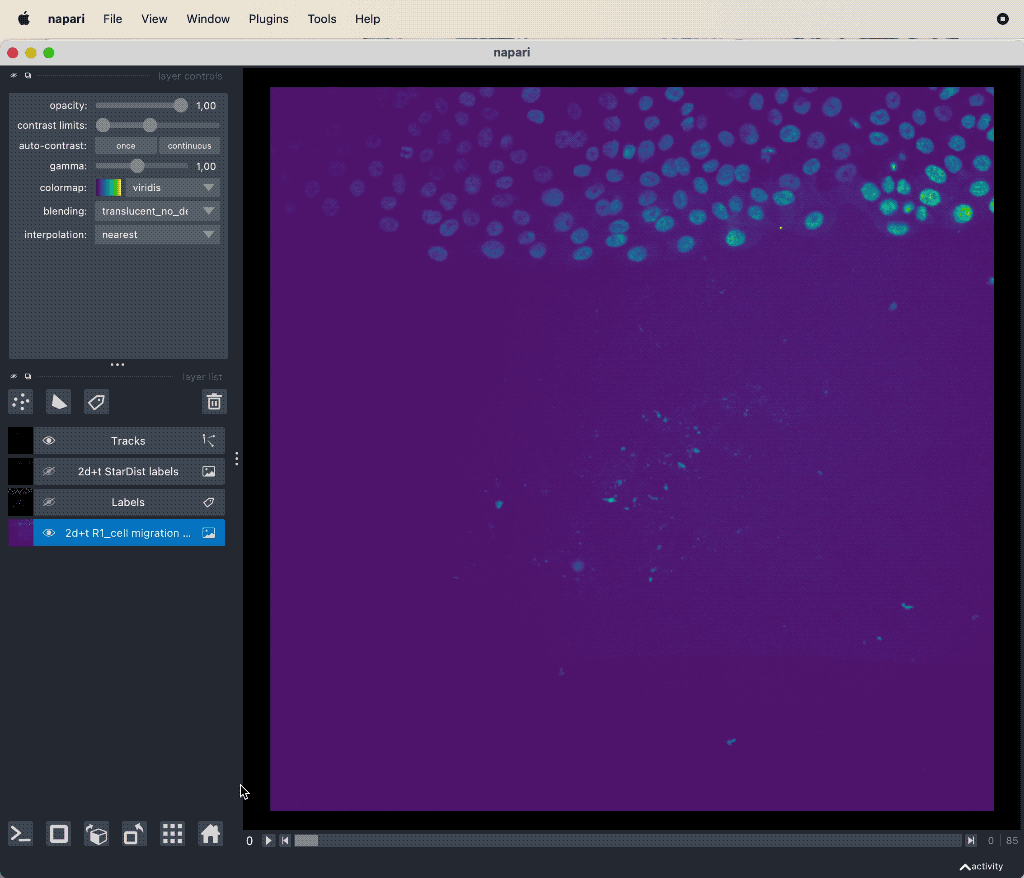 Track cell migration with Napari via the laptrack pluginꜛ.
Track cell migration with Napari via the laptrack pluginꜛ.
Why choose Napari?
Napariꜛ is a powerful free and open-source software (FOSS)ꜛ tool that has captivated me with its versatility in bioimage analysis. Written in on Python, its user-friendly interface and extensive functionality make it an ideal choice for visualizing, analyzing, and annotating biological images. With Napari, you can explore and manipulate small to large datasets in real-time with ease, while having access to community-driven, latest state-of-art image-processing tools through addable pluginsꜛ. Being part of an active community also means regular updates, bug fixes, and exciting new features.
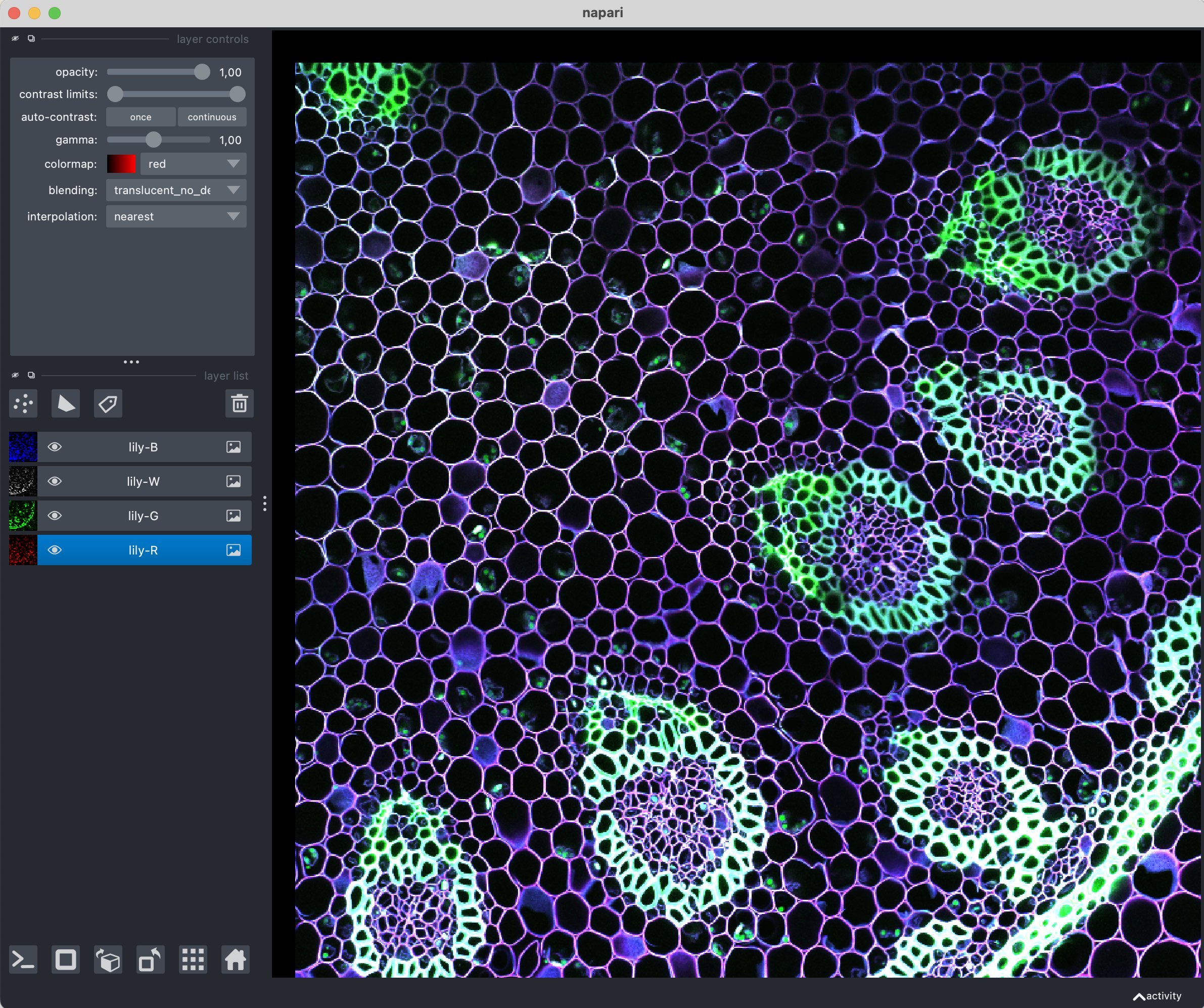 Napari contains a broad set of sample image, which already come pre-installed. The course material contains further freely available bioimage sample for you to explore.
Napari contains a broad set of sample image, which already come pre-installed. The course material contains further freely available bioimage sample for you to explore.
Sharing and accessibility
I firmly believe that knowledge should be freely accessible to all. Therefore, the entire material is available for everyone to use. Whether you are an individual learner, an educator, or a researcher seeking efficient tools for your lab, just explore, please share, and adapt the content to suit your needs. If you find any errors or have suggestions for improvement, please let me know in the comment section below, via email, or on Mastodonꜛ. I will be happy to hear from you.
comments