Image denoising and background subtraction in Napari
Two common preprocessing steps in bioimage analysis are image denoising and background subtraction. Both steps aim to improve the quality of the image and to facilitate subsequent image analysis tasks. In the following, both steps are briefly described and demonstrated in Napari.
If you haven’t installed Napari yet, please do so before proceeding with this tutorial. To practice this tutorial, install the following package(s) in your activated Napari conda environment:
mamba install -y napari-assistant napari-simpleitk-image-processing
What is image denoising?
Image denoising refers to the process of reducing or removing unwanted noise from digital images. Noise in images can arise from various sources, such as sensor limitations, transmission interference, or environmental factors. The presence of noise can degrade image quality, blur details, and impact the accuracy of subsequent image analysis tasks. Image denoising techniques aim to restore the original content of the image by suppressing or eliminating the noise while preserving important image features and details. Standard image denoising techniques include spatial filtering, frequency domain filtering, and wavelet-based filtering. In recent time, many deep learning-based denoising methods have been proposed. In this tutorial we will focus on spatial filtering. An overview of deep-learning based denoising methods can be found here.

 Example of noise reduction using spatial filtering techniques. Common filters applied for image denoising are the median filter, Gaussian filter, and mean filter. The code to generate these plots can be found in the GitHub repository of this course.
Example of noise reduction using spatial filtering techniques. Common filters applied for image denoising are the median filter, Gaussian filter, and mean filter. The code to generate these plots can be found in the GitHub repository of this course.
Spatial filtering for noise reduction is based on the convolutionꜛ of the image $f(x,y)$ with a filter kernel $w$, resulting in the “filtered”, i.e. convolved image $g(x,y) = w \ast f(x,y)$ ($\ast$ stands for the convolution). The filter kernel $w$ is a small matrix that is applied to each pixel of the image $f$. Filtering, i.e., convolution is the process of adding each element of the image $f$ to its local neighbors, weighted by the kernel $w$:
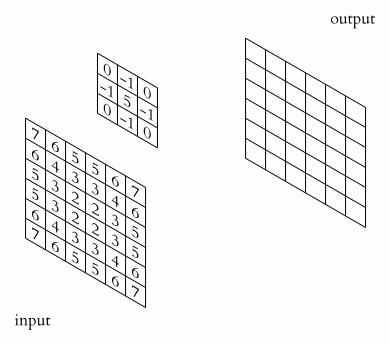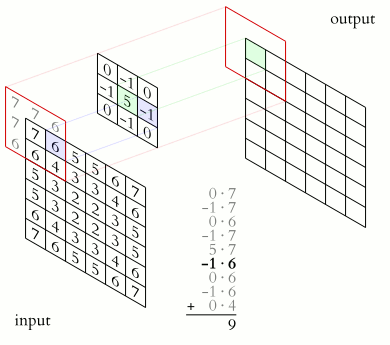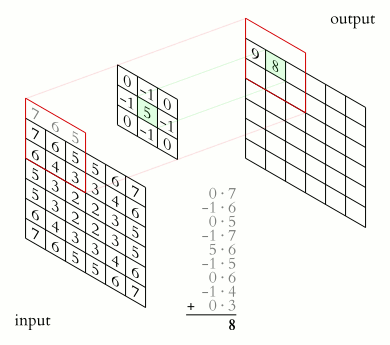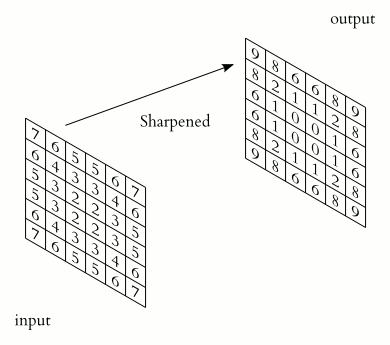 Animation of 2D convolution. The input is convolved with a 3x3 filter kernel, which weights each input pixel, resulting into a new output, i.e., convolved or “filtered” image. Source: Wikimediaꜛ (CC BY-SA 3.0 license)
Animation of 2D convolution. The input is convolved with a 3x3 filter kernel, which weights each input pixel, resulting into a new output, i.e., convolved or “filtered” image. Source: Wikimediaꜛ (CC BY-SA 3.0 license)
The type of filtering is determined by the type, i.e., by the values of the filter kernel. Here are some common 3x3 kernels, on a sample image without noise:
The median filter is a non-linear filter and is not represented by a kernel. Instead, the median filter replaces each pixel’s value with the median value of its neighboring pixels. To calculate the median filter, you slide a window over the image and replace the center pixel with the median value of the pixel values within the window. The median value is the middle value when all the pixel values in the window are sorted.
The common filters kernels for image denoising are the median, Gaussian, and mean filter.
What is background subtraction?
Background subtraction refers to the process of removing the background from an image. The background is defined as the part of the image that does not contain the object of interest, i.e., relevant information. Background subtraction is often used to facilitate subsequent image analysis tasks, such as object detection, segmentation, or tracking. It can be performed in a variety of ways, including thresholding, morphological operations, and machine learning-based methods.
 White top-hat (middle) and black top-hat (right) background subtraction method applied on a sample image (left). The code to generate these plots can be found in the GitHub repository of this course.
White top-hat (middle) and black top-hat (right) background subtraction method applied on a sample image (left). The code to generate these plots can be found in the GitHub repository of this course.
Examples for other background subtraction methods are:
- Top-hat background subtraction: Top-hat background subtraction is a morphological operation used for isolating small details or structures in an image that are smaller than a specified structuring element. White top-hat subtraction involves subtracting the morphological opening from the original image, highlighting bright structures or background variations. Black top-hat subtraction subtracts the original image from the morphological closing, emphasizing dark structures against a brighter background. These techniques are useful for detecting small foreground objects or correcting uneven illumination in image analysis tasks.
- Rolling Ball Algorithm: The rolling ball algorithm is a morphological operation used for background subtraction. It involves the rolling of a ball-like structuring element over the image, where the size of the ball is defined by a user-defined radius. At each position, the algorithm computes the minimum intensity value within the ball’s footprint, which represents the local background. The original image is then subtracted by this estimated background to obtain the background-subtracted result.
- Gaussian Mixture Model (GMM) Subtraction: GMM is a probabilistic model that represents the image as a mixture of Gaussian distributions. The background is modeled as one or more Gaussian components, and foreground objects are detected by comparing the pixel values with the GMM.
- Frame Differencing: In frame differencing, consecutive frames in a video sequence are subtracted to detect changes. The static background is estimated by averaging or selecting a representative frame, and the differences between subsequent frames indicate moving objects.
- Robust Principal Component Analysis (RPCA): RPCA decomposes the image into a low-rank background component and a sparse foreground component. By separating the image into these components, the background can be extracted.
- Adaptive Background Subtraction: This technique adapts the background model over time to handle gradual changes in illumination or scene dynamics. It updates the background model based on statistical measures or adaptive learning algorithms.
- Eigenbackground Subtraction: Eigenbackground assumes that the background variations can be represented by a subspace. The background is modeled using principal component analysis (PCA) to capture the dominant variations, and deviations from the background model indicate foreground objects.
- Thresholding: Thresholding is a technique where pixels in an image are divided into two groups based on a specific threshold value. This separation allows the identification and separation of foreground objects from the background. Thresholding can be performed using a global threshold value or an adaptive threshold value that varies across the image.
Image denoising and background subtraction with Napari
In Napari, functions for both image denoising and background subtraction are provided by the napari-simpleitk-image-processingꜛ plugin. This plugin not only contains functions for two mentioned techniques, but the also offer a broad set of various image processing functions in general.
Applying image denoising and background subtraction very simple. You can access them via Tools menu and then choose a function either from Filtering / noise removal to apply a noise reduction method or from Filtering / background removal to apply a top-hat background subtraction:
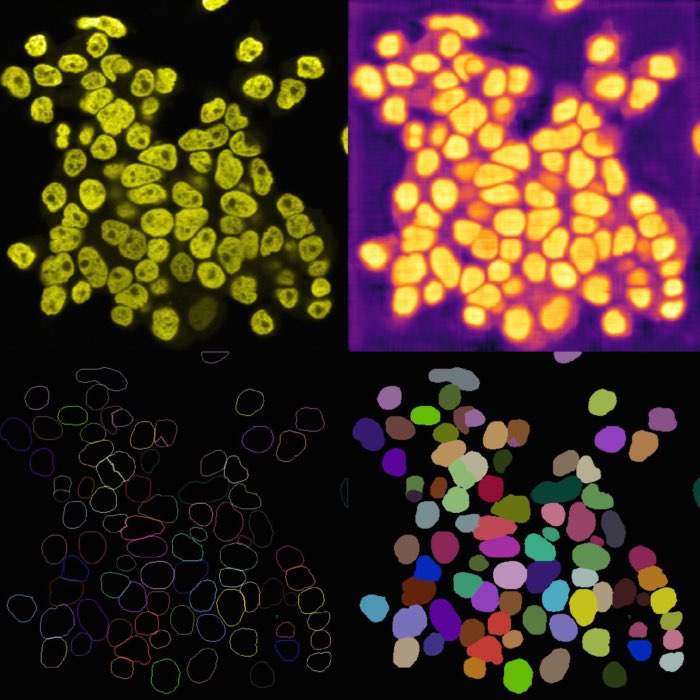
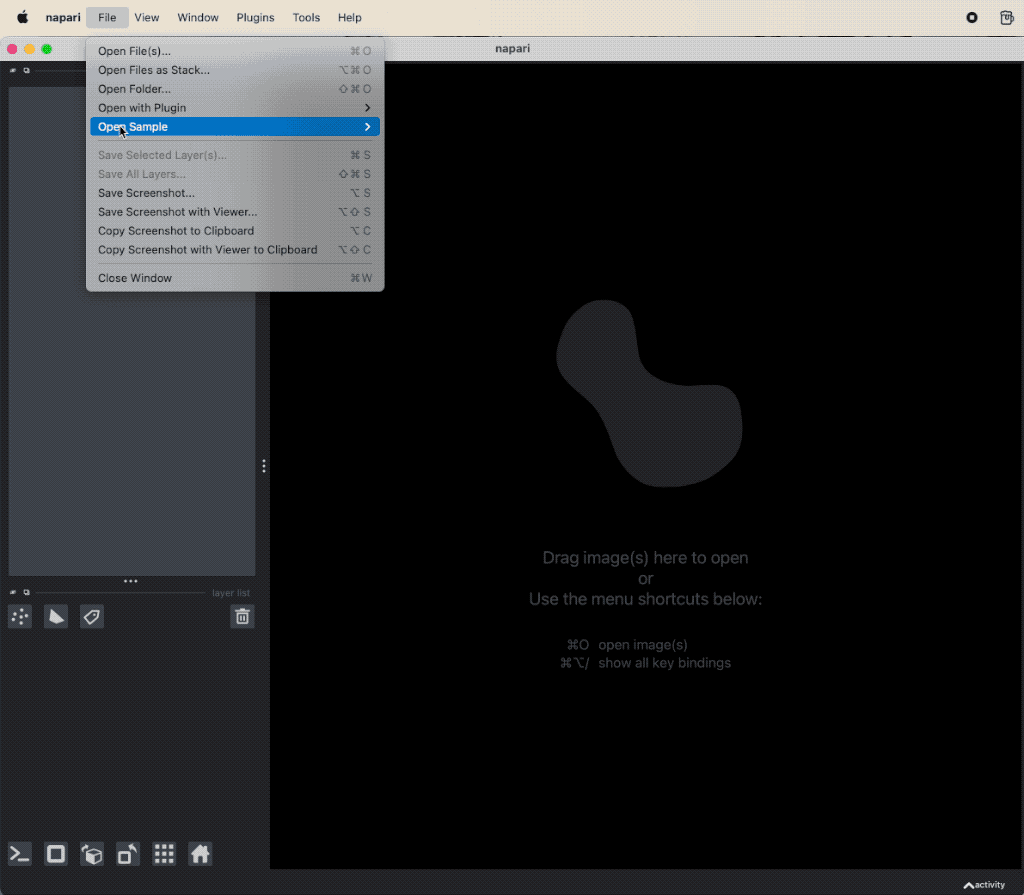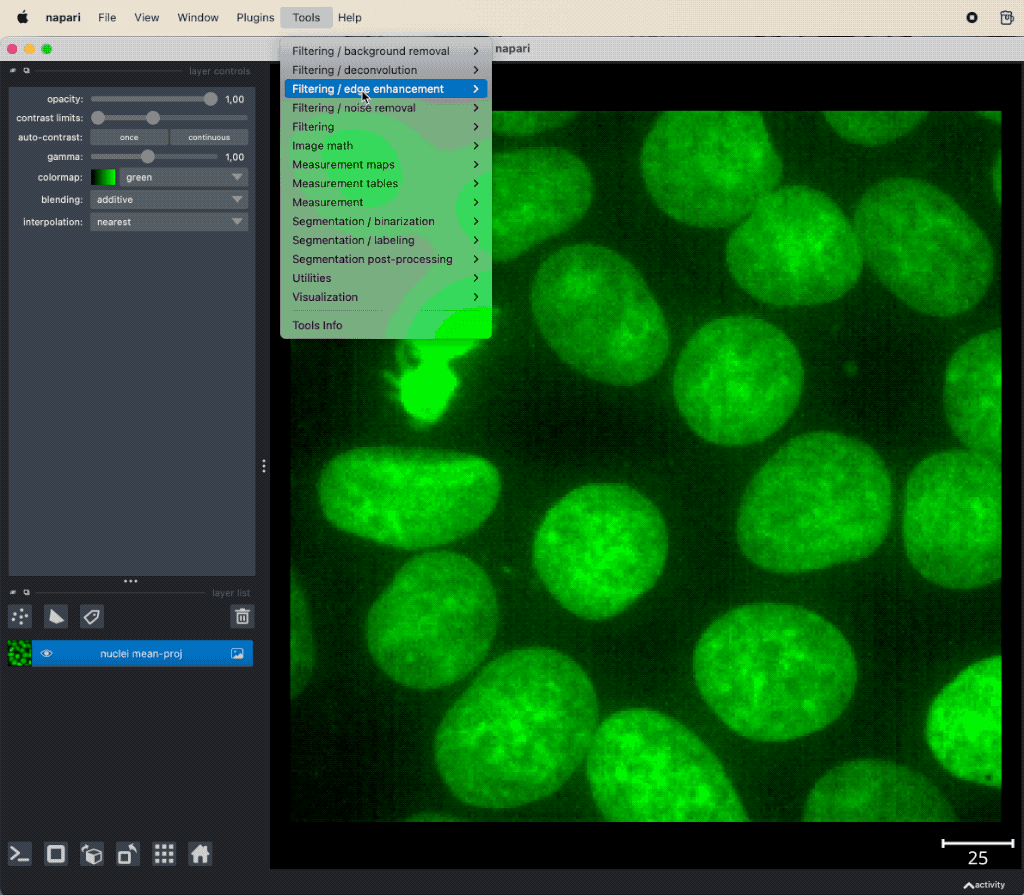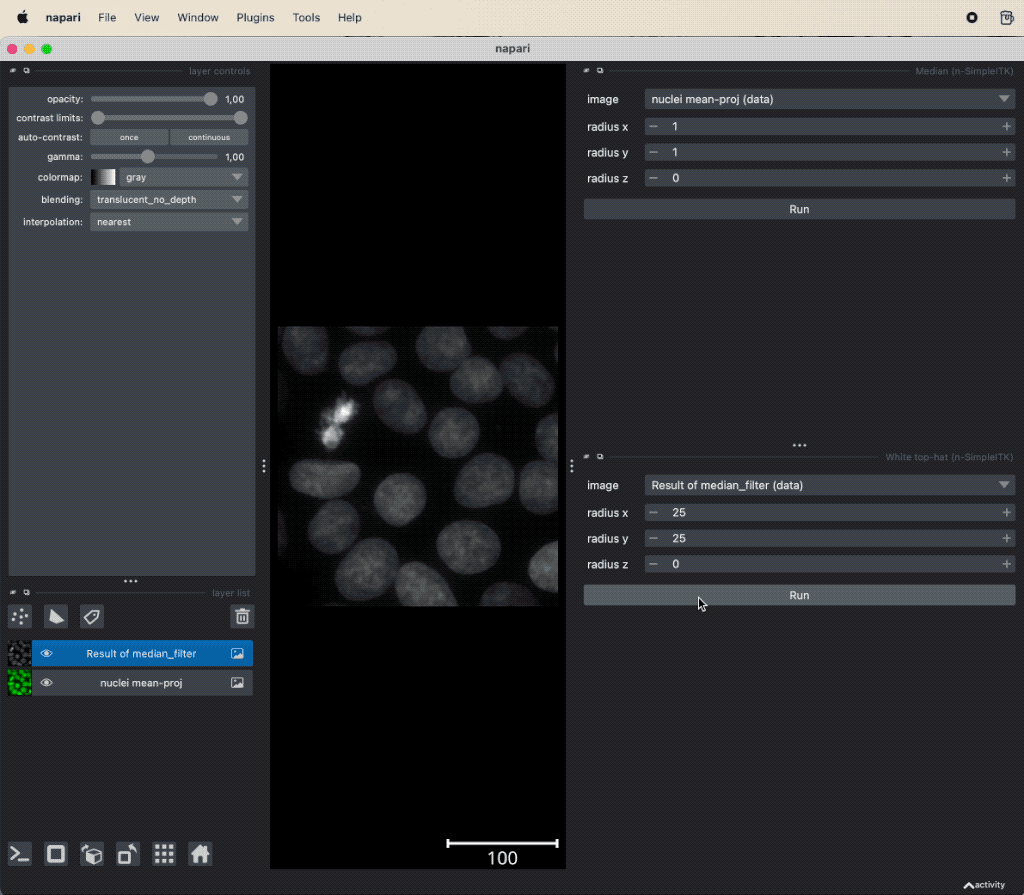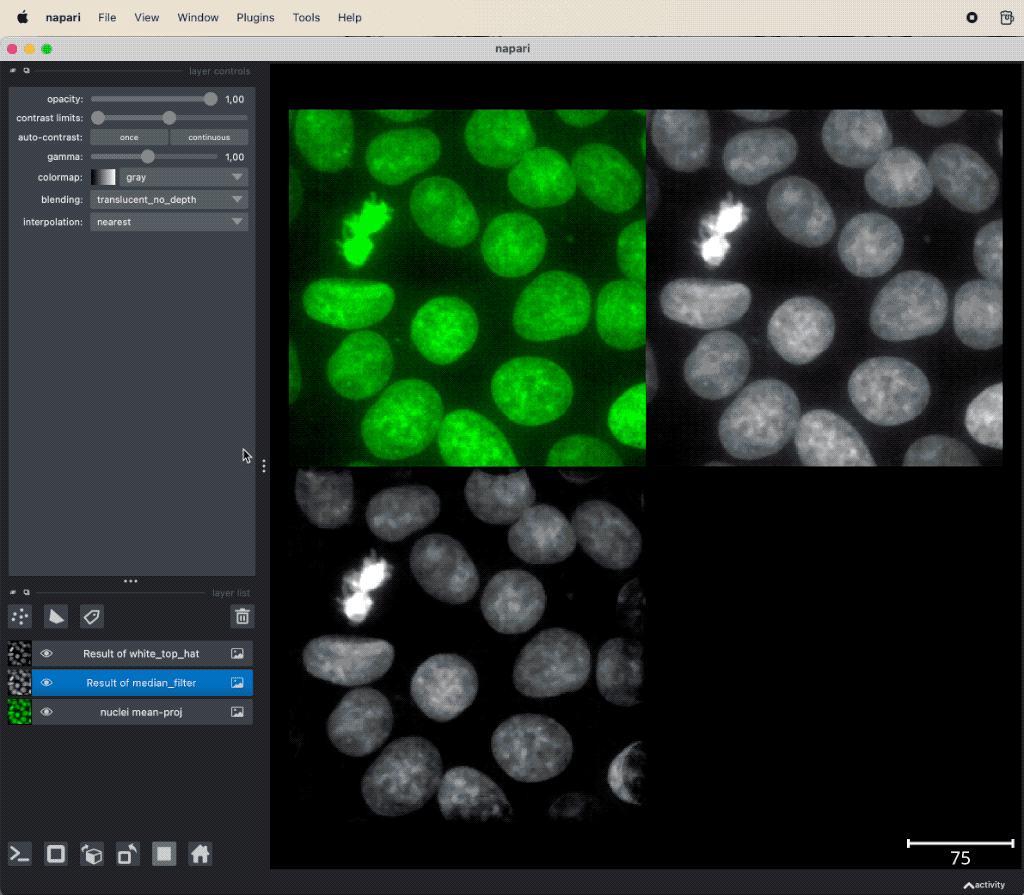 Example of image denoising and background subtraction applied to the projection of nuclei layer of the “Cells (3D+2Ch)” sample image. Both plugins come along with the napari-simpleitk-image-processingꜛ plugin
Example of image denoising and background subtraction applied to the projection of nuclei layer of the “Cells (3D+2Ch)” sample image. Both plugins come along with the napari-simpleitk-image-processingꜛ plugin
Exercise
Please apply image denoising and background subtraction to the “nuclei_2D_projection_snp_noisy” and “nuclei_2D_projection_gaussian_noisy” sample images. Please try different denoising functions and settings. What do you notice?

_filtered.png)
_filtered.png)
_filtered.png)
_filtered.png)
_filtered.png)
_filtered.png)