NEST simulator – A powerful tool for simulating large-scale spiking neural networks
The NEST (NEural Simulation Tool) simulator is a powerful open source software tool designed for simulating large-scale networks of spiking neurons (SNN). It has become an essential instrument in the field of computational neuroscience, providing the capability to model, simulate, and analyze the complex dynamics of neuronal systems. And it comes with a user-friendly Python interface, facilitating the construction of neuronal networks with minimal effort.
 NEST logo. Source: github.com/nestꜛ
NEST logo. Source: github.com/nestꜛ
Development history
NEST development began in 1993 by Markus Diesmann and Marc-Oliver Gewaltig at the Ruhr University Bochum, Germany, and the Weizmann Institute of Science in Rehovot, Israel. Initially called SYNOD and utilizing a stack-based simulation language named SLI, the software was renamed NEST in 2001. Until 2004, NEST was exclusively developed and used by the founding members of the NEST Initiativeꜛ, with the first public release appearing in the summer of that year. Since then, NEST has been regularly updated, typically releasing new versions once or twice a year.
In 2007, NEST introduced hybrid parallelism using POSIX threadsꜛ and MPIꜛ, both enabling shared-memory and distributed-memory parallelism. Also the stack-based SLI language was largely replaced by a modern Python interface in 2008 (although SLI remains in use internally). Simultaneously, the simulator-independent specification language PyNNꜛ was developed to support NEST. PyNN is a common interface for neuronal network simulators, allowing users to write code that can run on multiple simulators such as NEURONꜛ, Brianꜛ – and NEST – without modification. In 2012, the NEST Initiative transitioned from the proprietary NEST License to the GNU GPL V2 or later.
Architecture, design, and philosophy
NEST is built to handle networks with an extensive number of neurons and synapses, ensuring efficient memory usage and computational speed. Designed to mimic the logic of electrophysiological experiments within a computer environment, NEST requires the users to explicitly define the neural system under investigation, providing all necessary tools for this purpose.
NEST supports a wide range of experimental paradigms, including setups with multiple neuron populations, complex connectivity patterns, and various input stimuli. Different neuron and synapse models can coexist, allowing for diverse neuronal connections.
To manipulate or observe network dynamics, virtual recording devices can be defined within the simulation. These devices simulate instruments used in real experiments for measurement and stimulation, recording data either to memory or to a file. NEST’s extensibility allows the addition of new models for neurons, synapses, and devices as needed.
In summary, the core design principles of NEST are scalability, flexibility, and precision:
Scalability – NEST employs advanced algorithms and data structures to manage large-scale simulations efficiently. It leverages parallel computing architectures, such as multi-core processors and distributed computing systems, enhancing performance and making it suitable for networks with millions of neurons and billions of synapses.
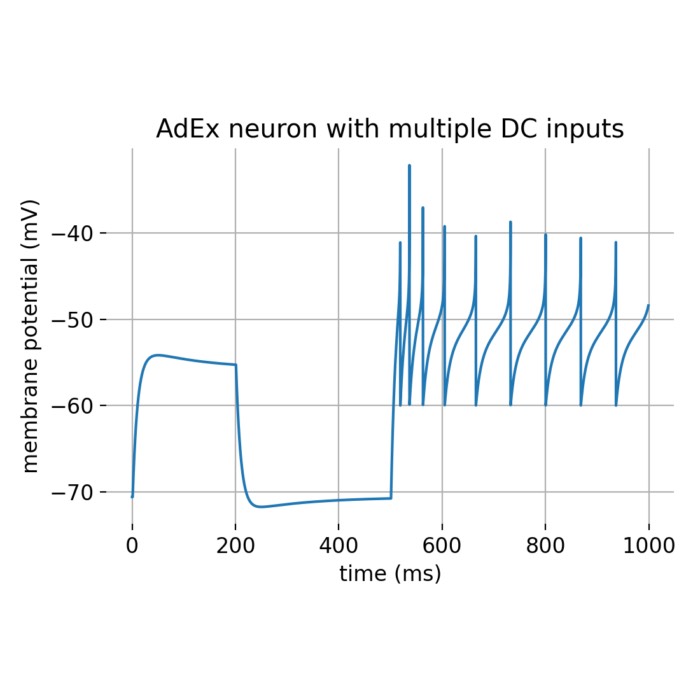
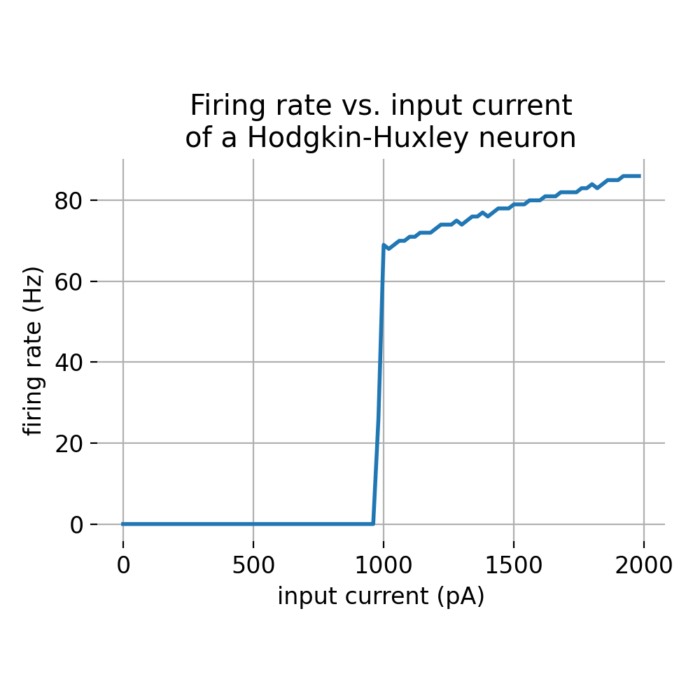
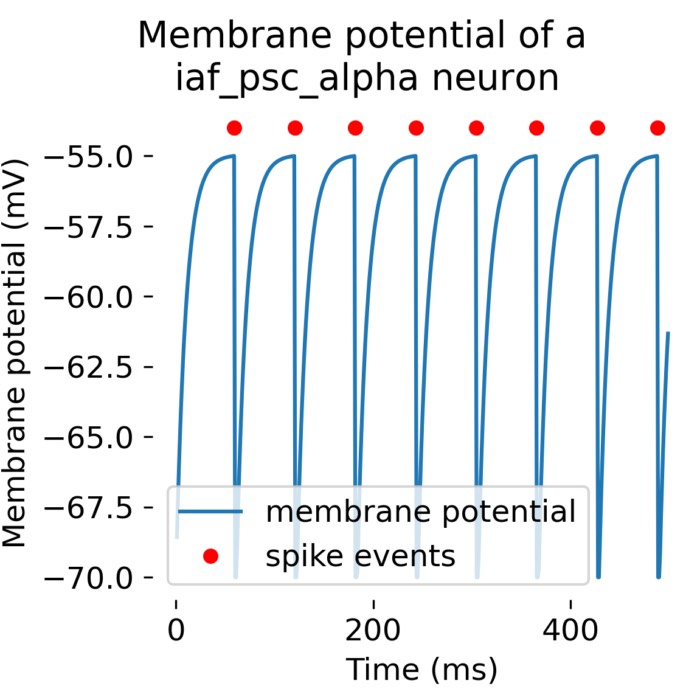
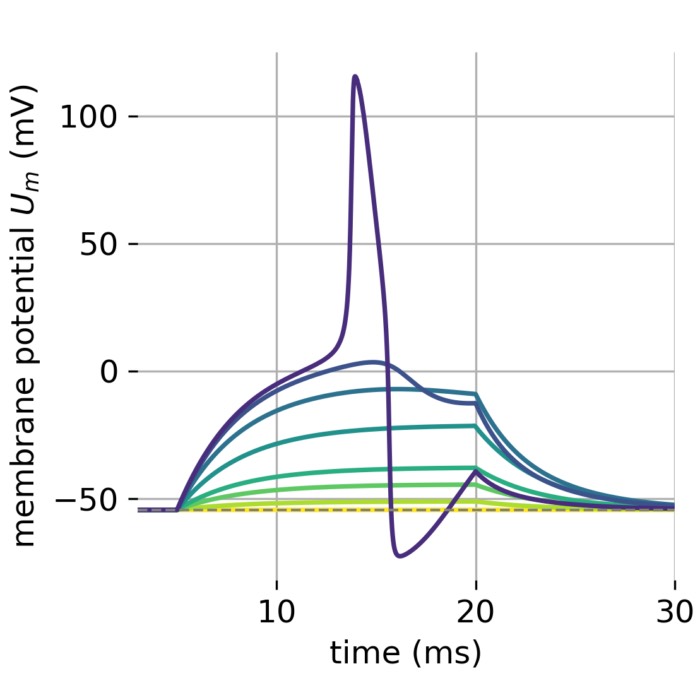
Flexibility – The simulator supports various neuron and synapse models, including integrate-and-fire neurons, Hodgkin-Huxley-type neurons, and plastic synapses. Users can extend functionality by defining custom models and incorporating them into the simulation environment, making NEST versatile for diverse research applications.
Precision – NEST ensures high numerical accuracy through precise time integration methods and accurate representation of neuronal dynamics, critical for capturing intricate neural behavior and producing reliable simulation results.
The primary user interface for NEST is PyNEST, a Python package that provides a high-level interface to the simulator. PyNEST simplifies the construction of neuronal networks, simulation control, and data analysis, and enables to focus on experimental design and data interpretation rather than low-level programming tasks.
Key features
NEST offers a wide range of features that make it a versatile tool for neuroscientific research:
Modeling – Users can construct complex neuronal networks using a high-level language interface, primarily Python. NEST provides a rich library of pre-defined neuron and synapse modelsꜛ, allowing users to create detailed simulations with minimal effort.
Simulation control – NEST offers extensive control over simulation parameters, including the ability to specify simulation duration, time resolution, and input stimuli. This flexibility enables users to tailor simulations to their specific experimental needs.
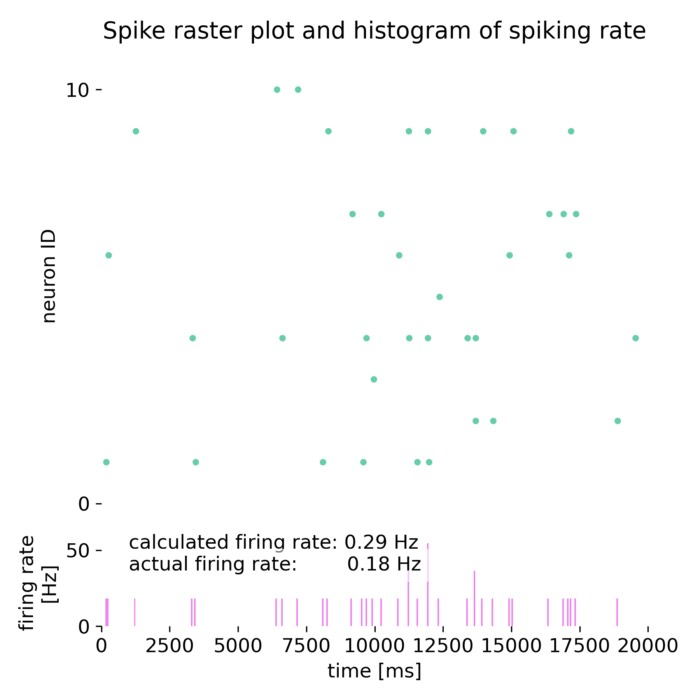
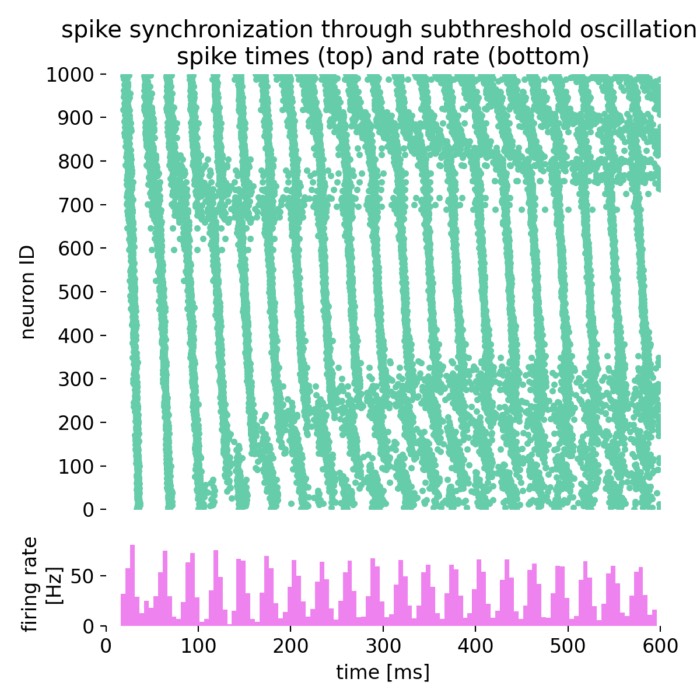
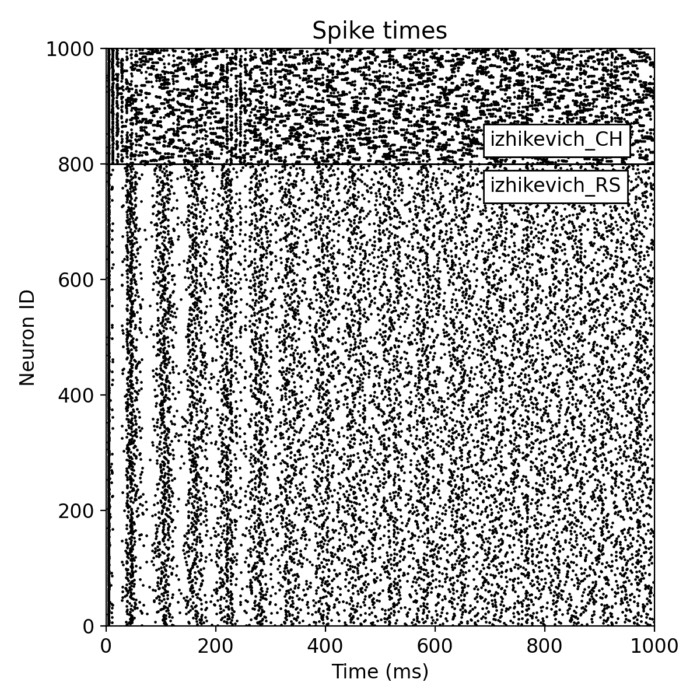
Analysis tools – The simulator includes built-in tools for analyzing simulation data, such as spike train analysis, raster plots, and firing rate calculations. Additionally, NEST supports integration with external data analysis tools, like NumPy and SciPy, for advanced data processing.
Parallel computing – To handle large-scale simulations, NEST is designed to run efficiently on parallel computing infrastructuresꜛ. It supports both shared-memory and distributed-memory systems, enabling users to utilize high-performance computing resources effectively.
Extensibility – NEST’s modular architecture allows users to extend its functionality by adding custom neuron and synapse models, as well as new simulation and analysis tools. This extensibility is facilitated by a well-documented APIꜛ and a supportive user communityꜛ.
Applications in neuroscience
NEST has been instrumental in advancing the understanding of neural dynamics and network behavior. Its applications span various domains within neuroscience:
Neural coding – NEST can be used to study how neurons encode and process information. By simulating different coding schemes, such as rate coding and temporal coding, one can investigate the principles underlying neural representation and computation.
Network dynamics – NEST allows the exploration of dynamic phenomena in neural networks, such as oscillations, synchrony, and propagation of activity. These studies provide insights into the mechanisms of information processing and communication in the brain.
Plasticity and learning – The simulator is employed to model synaptic plasticity mechanisms, such as Hebbian learning and spike-timing-dependent plasticity (STDP). These models help elucidate how learning and memory processes are implemented at the synaptic level.
Brain disorders – By simulating pathological conditions, like epilepsy and Parkinson’s disease, NEST contributes to the understanding of disease mechanisms and the development of therapeutic interventions. It enables the testing of hypotheses about disease progression and the effects of potential treatments.
Conclusion
The NEST simulator stands as a cornerstone in computational neuroscience, providing a robust and versatile platform for simulating and analyzing large-scale neuronal networks. Its scalable architecture, flexible modeling capabilities, and precise simulation methods make it invaluable for studying brain function, neural dynamics, and neurological disorders. By enabling detailed investigations into the complex interplay of neurons and synapses, NEST facilitates the exploration of fundamental questions in neuroscience. With its user-friendly Python interface and extensive feature set, NEST enables this research to be conducted efficiently and effectively, from small-scale simulations on a laptop to large-scale models on high-performance computing clusters.
In my next posts, we will explore these capabilities further with short step-by-step examples, demonstrating how to set up and run simulations in NEST and analyze simulation results. Stay tuned.
 NEST terminal output.
NEST terminal output.
References and useful links
Publications
- Gewaltig, M.-O., & Diesmann, M., NEST (NEural Simulation Tool), 2007 Scholarpedia, 2(4), 1430, doi: 10.4249/scholarpedia.1430ꜛ
- Eppler, J. M., Helias, M., Muller, E., Diesmann, M., & Gewaltig, M.-O., PyNEST: a convenient interface to the NEST simulator, 2009, Frontiers in Neuroinformatics, 2, 12, doi: 10.3389/neuro.11.012.2008ꜛ
- Davison, A. P.; Brüderle, D.; Eppler, J.; Kremkow, J.; Muller, E.; Pecevski, D.; Perrinet, L.; Yger, P. PyNN: A Common Interface for Neuronal Network Simulators, 2009, Frontiers in Neuroinformatics. 2: 11. doi: 10.3389/neuro.11.011.2008ꜛ
- Plesser, Hans E.; Eppler, Jochen M.; Morrison, Abigail; Diesmann, Markus; Gewaltig, Marc-Oliver, Efficient Parallel Simulation of Large-Scale Neuronal Networks on Clusters of Multiprocessor Computers, 2007, Euro-Par 2007 Parallel Processing, Lecture Notes in Computer Science. Vol. 4641. pp. 672–681. doi: 10.1007/978-3-540-74466-5_71ꜛ
- Morrison, Abigail; Straube, Sirko; Plesser, Hans Ekkehard; Diesmann, Markus, Exact Subthreshold Integration with Continuous Spike Times in Discrete-Time Neural Network Simulations, 2007, Neural Computation. 19 (1): 47–79. doi: 10.1162/neco.2007.19.1.47ꜛ
- Jordan, J., Ippen, T., Helias, M., Kitayama, I., Sato, M., Igarashi, J., … & Diesmann, M., Extremely scalable spiking neuronal network simulation code: From laptops to exascale computers, 2018, Frontiers in Neuroinformatics, 12, 2, doi: 10.3389/fninf.2018.00002ꜛ
- NEST - A brain simulator, Bernstein Network. 2012-07-11, via YouTubeꜛ
- Wikipedia article on the NEST simulatorꜛ
comments